



|
Docking of Select Cinnamon Components Suggest Potential for Sirt-1 Activation Similar to Resveratrol
Martin Brenneman, Amy Stockert*, David Kinder, Tarek Mahfouz |
|
Raabe College of Pharmacy, Department of Pharmaceutical and Biomedical Sciences, Ohio Northern University, USA. |
ABSTRACT
Objectives: Cinnamon has anti-hyperglycemic effects by multiple sources. However, it is not known which component in the cinnamon extract is responsible for this effect nor is it known what the protein targets are. Sirtuin 1 (Sirt-1) deacetylase is involved in chromatin remodeling and control of gene expression. Since many proteins in the insulin signaling pathway are regulated by acetylation, Sirt-1 is a potential player in anti-hyperglycemic mechanisms. Multiple cinnamon components bear high structural similarity to resveratrol, a plant polyphenol whose anti-hyperglycemic effect has been attributed to direct activation of Sirt-1. This docking study was initiated to investigate the binding potential of those components to Sirt-1 to guide further experimental research into the anti-hyperglycemic mechanisms of cinnamon and to develop a method for screening potential Sirt-1 activators. AutoDock Vina was used to dock resveratrol and the major components in the cinnamon extract to Sirt-1. The docking behavior and hydrogen-bonding pattern of each cinnamon component were compared to those of resveratrol to identify similarities. The polyphenol quercetin displayed the most similar behavior to resveratrol whereas cinnamic acid and its derivatives were the most dissimilar. These results suggest that quercetin most likely is the ingredient in cinnamon to activate Sirt-1, potentially leading to the anti-hyperglycemic effects.
Key Words: SIRT-1 activators; sirtuin, docking; energy metabolism; resveratrol; cinnamon; quercetin; antihyperglycemic herbs; medicinal herbs.
INTRODUCTION
Health promotion is of the responsibility of individuals and can lead to higher levels of health [1-4]. The medicinal properties of cinnamon have been recognized for years ranging from use for gastrointestinal discomfort to blood sugar control [5]. In a previous study, we have shown that cinnamon extract was effective at reducing blood glucose [6]. Similarly, other studies have demonstrated anti-hyperglycemic effects as well as additional health benefits of individual components in the cinnamon extract [7, 8]. However, the details of the anti-hyperglycemic mechanism of cinnamon are not completely understood.
In mouse 3T3-L1 preadipocytes, cinnamon extracts have been shown to increase expression of GLUT 4 [9], the primary glucose transporter in muscle and adipose tissue, but the mechanism by which induction occurs has not been elucidated. Clinical studies suggest cinnamon extract affects both fasting and postprandial glucose levels, indicating that cinnamon affects the liver as well as muscle and adipose tissues. Thus, the protein target, or targets, involved must be present in at least three tissue types. Isoforms of Sirt enzymes exist in these tissues and, therefore, could be a potential target for the components from the cinnamon extract [6].
A literature survey yielded several other natural products with biological effects similar to cinnamon. Examples include aspalathin from Rooibus [10] caffeic acid from coffee [8], fraxetin from the Chinese herb Cortex fraxini [11], ferulic acid [12], and resveratrol [13]. Of those, resveratrol is the best characterized and well-studied in the isolated pure state. Resveratrol has been shown to have anti-hyperglycemic effects [14] similar to cinnamon. Importantly, its mechanism of anti-hyperglycemia and other pharmacological effects has been attributed to its activation of human Sirt-1 deacetylase. Sirt-1 is an enzyme with multiple tissue isoforms found in the liver, muscle, the adipose, and other tissues [15, 16]. Despite the controversy surrounding resveratrol-Sirt-1 interactions [17, 18], resveratrol is a stimulator in sirtuin-dependent processes extensively [19-22]. Sirt-1 has been co-crystalized with a peptide substrate and resveratrol bound in the active site indicating the direct interaction between resveratrol and Sirt-1 (PDB code 5BTR [23]).
Sirtuin enzymes make up a group of structurally and functionally similar proteins that have demonstrated functional roles in chromatin remodeling and metabolism regulation, especially carbohydrate metabolism [24]. These biological functions of the sirtuins suggest they might be a potential target for the cinnamon components. The major ingredients in the cinnamon extract are the cinnamic acid and its derivatives (cinnamyl alcohol and cinnamaldehyde) in addition to glycosidic and polyphenolic molecules [5], Figure 1. Many of those molecules share structural similarity to resveratrol, especially the polyphenols, Figures 1(a) and 1(b). It is these structural similarities that lead us to hypothesize that these components of cinnamon are activators of Sirt-1 and contribute to the anti-hyperglycemic effects observed in the clinical use of cinnamon, similar to resveratrol.
The goal of this docking study is to guide further research aimed at identifying components of the cinnamon extract with the potential to achieve anti-hyperglycemic effects and other health benefits by affecting Sirt-1 substrate interactions.
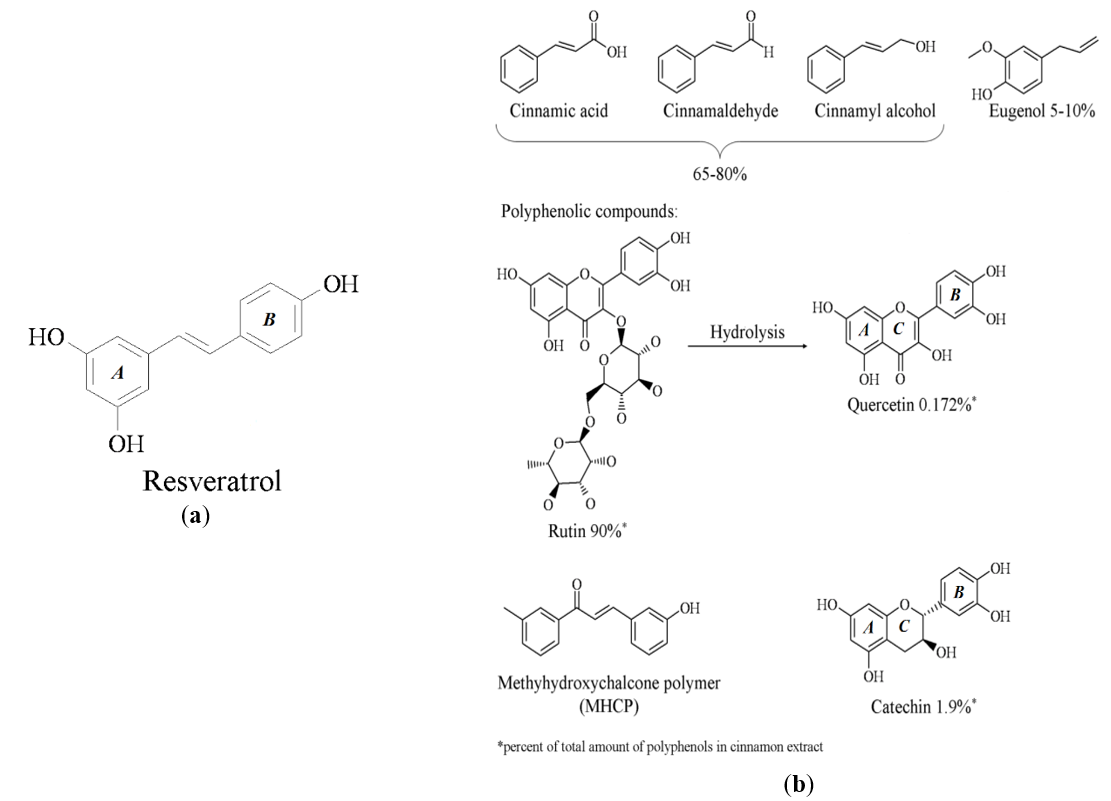
Figure 1. (a) Chemical structure of resveratrol; (b) Chemical structures of major components of the cinnamon extract.
RESULTS AND DISCUSSION
Docking of resveratrol overlaid with the crystal structure of resveratrol validates the docking model and supports the cooperative binding model
Crystal structure 5BTR shows three resveratrol molecules that each bind in a separate subsite within the large active site of the Sirt-1 deacetylase. Those three subsites were labeled S1, S2, and S3, Figure 2(a). Those three resveratrol molecules enhance the deacetylase activity of Sirt-1 by further stabilizing the substrate peptide in the active site by mediating several hydrogen bonding (H-bonding) interactions between active site residues and the substrate peptide [9], Figure 2(b). These interactions are summarized here. In S1: one of the two OH groups on ring A (see ring numbering in Figure 1) of resveratrol forms an H-bond with the side chain of E230. The other OH on the same ring H-bonds to the substrate’s K3 residue’s backbone carbonyl oxygen. The sole OH group on resveratrol’s ring B H-bonds to Sirt-1’s backbone amide nitrogen of residue P211. In S2: only one of the OH groups on ring A H-bonds to the side chain of Sirt-1’s D298 while the sole OH on ring B H-bonds to the substrate’s backbone carbonyl oxygen of residue R1 and each of Sirt-1’s side chains of Q222 and N226. In S3: one of the OH groups on ring A H-bonds to Sirt-1’s side chain of residue D298 while the other OH of the same ring H-bonds to Sirt-1’s side chain of D292. The sole OH group on ring B H-bonds to the oxygen of the fluorescent group (FDL) of the peptide substrate. It is worth noting here that of the three resveratrol in the Sirt-1 active site, only the one in S3 forms direct contact with the FDL fluorescent group as explained in the previous sentence.
In addition to hydrogen bonding interactions, hydrophobic interactions likely contribute to the stabilizing effect of resveratrol, primarily in S1 and S3 with the FDL group. Logically, other molecules forming similar hydrogen bonding patterns or hydrophobic interactions within this network of sites should also stabilize the substrate in the active site and function as a Sirt-1 activator. For this reason, the different components of cinnamon extract were docked into the Sirt-1 binding site.
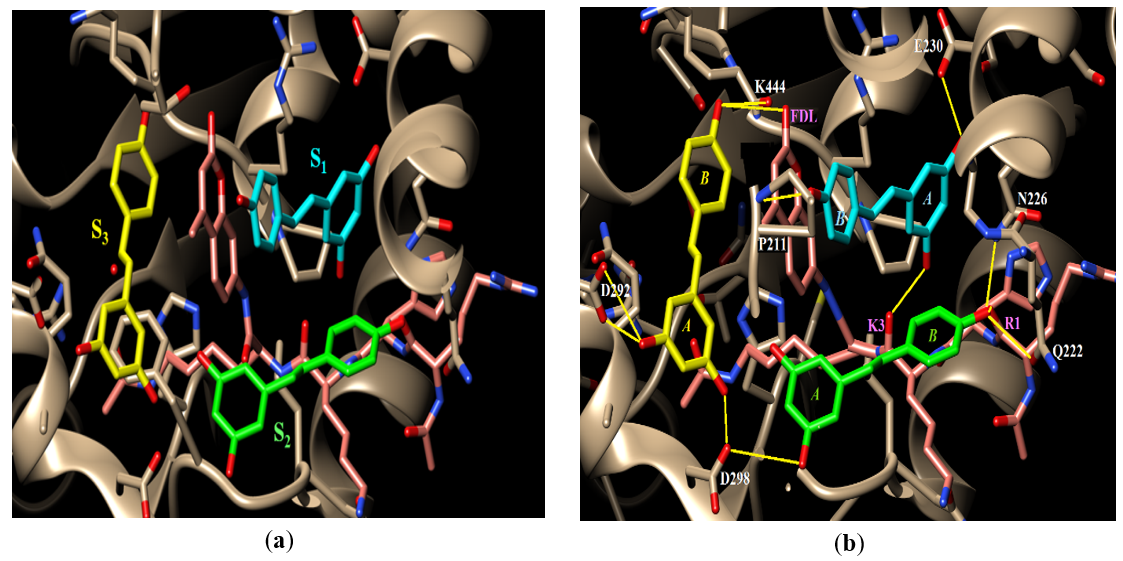
Figure 2. (a) The active site of Sirt-1 in structure 5BTR. The P53 substrate peptide is shown in pink, the first resveratrol molecule is shown in light blue, the second is shown in light green, and the third is shown in light yellow. (b) Hydrogen bonding interactions of the three resveratrol molecules in the binding site. Sirt-1 active site residues that H-bond to resveratrol are labeled in white whereas the substrate residues are labeled in pink. FDL is the fluorescent group of the substrate. H-bonds are indicated by yellow lines.
Our docking protocol involved keeping the substrate peptide in its crystal bound conformation unaltered but removing the three resveratrol molecules occupying the three subsites; S1, S2, and S3. A molecule of resveratrol was then built and docked into this evacuated binding site and the poses obtained were compared to the crystal bound conformations. Then, a molecule of each of the components reported to exist in the cinnamon extract [5] was docked separately and individually into the resveratrol-free Sirt-1 active site and the resulting docking poses and their scores were examined and compared to those observed for resveratrol in structure 5 BTR. Also, in examining the docking results and to differentiate between biologically relevant and irrelevant binding modes and to select poses for score reporting, only docking poses that met the following criteria were selected:
Although resveratrol is not a component of cinnamon extract, it was docked into the Sirt-1 active site for two reasons:
A resveratrol molecule was built as described in the Methods section and was then docked into the resveratrol-free active site of Sirt-1. Examination of the results of this first round of docking showed that resveratrol docked separately and individually into S1, S2, or S3. However, the docking pose that showed the most negative score among all poses across the three subsites was the one that docked in S1 with a score of -9.1, Figure 3a. It is important to also note that this docked pose also superimposed perfectly on the crystal conformation of resveratrol in S1 when overlaid on structure 5BTR, Figure 3A. The docking poses of resveratrol in S2 and S3 each had a score of -7.7, Figure 3A. This suggested that binding of resveratrol in S1 is most favored over binding in either S2 or S3.
To investigate whether binding of a resveratrol molecule in S1 might enhance the binding of resveratrol in S2, the coordinates of the -9.1 pose in S1 were added to the starting PDB file of the enzyme and the substrate peptide, and a new pdbqt file was generated with the same docking box and parameters as before and was used to dock the second resveratrol molecule in the second run of docking.
Upon examining the poses from the second run of docking (run 2 in Table 1), poses were observed in both S2 and S3 but the one with the most negative score was the pose in S2 with a score of -8.7; a 1 unit improvement from run 1, Table 1. Not only that but upon overlaying this pose on the crystal conformation of resveratrol in S2 in structure 5BTR it superimposed perfectly, Figure 3B. Similar to the previous step, To investigate whether binding of a resveratrol molecule in S2 might enhance the binding of resveratrol in S3, the coordinates of the -8.7 pose in S2 were added to the PDB file created in the previous run of the enzyme, the substrate peptide, and the resveratrol in S1 and a new pdbqt file was generated with the same docking box and parameters as before and was used to dock a third resveratrol molecule in the third run of docking (run 3, Table 1).
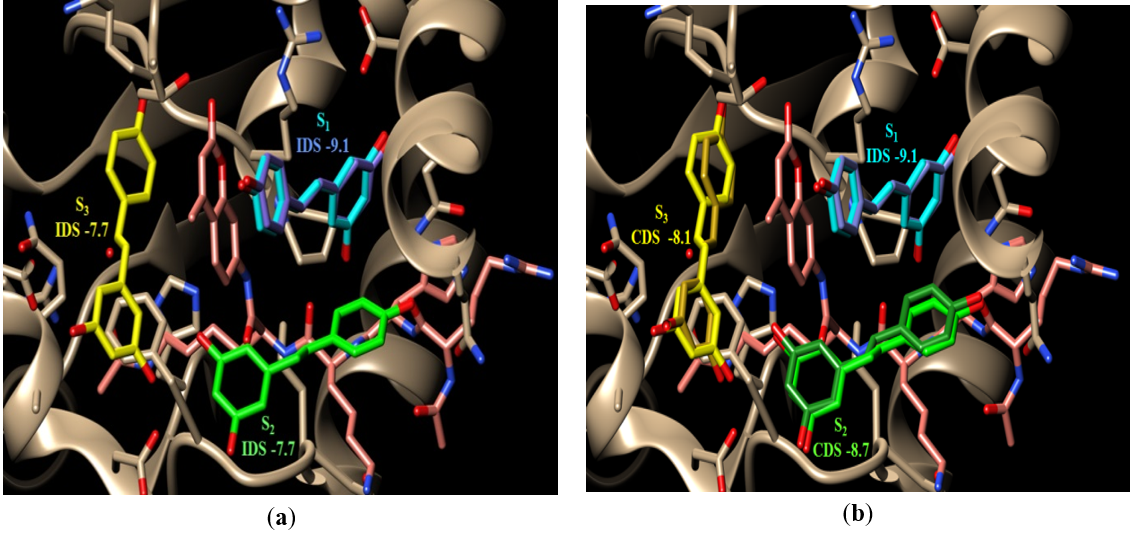
Figure 3. The binding site of Sirt-1 (PDB code 5BTR) with the three resveratrol molecules bound in the three subsites; S1 (cyan), S2 (green), and S3 (yellow). The P53 peptide is shown in pink; (a) the best scoring docking pose of resveratrol (darker blue) with its score are shown superimposed on the crystal bound conformation of resveratrol in S1. The docked poses in S2 and S3 are not shown. In all cases, the individual docking scores (IDS) for each subsite are shown underneath the subsite label. An IDS for a molecule is the docking score calculated by VINA for that molecule when it is docked into that subsite while the other two subsites are vacant (i.e. no molecules bound); (b) the docked resveratrol conformations (darker shade of the same color) superimposed on their corresponding crystal bound conformations (lighter shades). The cooperative docking score (CDS) is the docking scores calculated by VINA for a molecule docked in a subsite while at least one of the other two subsites is occupied by a docked molecule.
As expected, all resveratrol docked poses from run three were in S3 but the most negative scoring pose was -8.1; a 0.4 unit improvement from the same docking pose observed in S3 in run 1 when S1 and S2 were empty, Table 1. Again, upon overlaying this docking pose of run 3 on structure 5BTR it superimposed perfectly on the crystal conformation of resveratrol in S3, Figure 3b.
Table 1. Resveratrol Docking Scores
|
Run |
Subsite |
IDS |
Run |
Subsite |
CDS |
Run |
Subsite |
CDS |
|
1 |
1 |
-9.1 |
2 |
2 |
-8.7 1 |
3 |
3 |
-8.1 2 |
|
1 |
2 |
-7.7 1 |
|
|
|
|
|
|
|
1 |
3 |
-7.7 2 |
|
|
|
|
|
|
Individual docking scores (IDSs) and cooperative docking scores (CDS) of resveratrol in the three subsites S1, S2, and S3. Run 1 indicates the docking of 1 single molecule in the empty structure. Docking score of -9.1 indicated preference for S1 over S2 or S3. Run 2 indicated docking of a second molecule with S1 already occupied. 1demonstrated improvement in the docking score for resveratrol in S2 if resveratrol has already docked in S1. 2demonstrated improvement in the docking score for S3 if subsites 1 and 2 are already filled.
The best scoring docked poses of resveratrol overlaid their corresponding crystal bound conformations in all three subsites, thus validating our docking protocol. Also, the subsequent improvements in docking scores upon filling a previous subsite with resveratrol suggest a cooperative binding mode of resveratrol to Sirt-1 where one molecule facilitates the binding of the subsequent one.
Resveratrol docking allowed the establishment of a baseline from which to compare other docking results. Cinnamon extract components were selected for docking based on molecular structure and predicted percentage of the component.
Quercetin docking suggests cooperative binding as well as multiple binding modes.
Upon docking quercetin, it displayed docking behavior and scores that are very similar to those of resveratrol. First, quercetin binds most favorably in S1 with a docking score of -8.8 (that of resveratrol was -9.1, Table 2) and, more importantly, the quercetin docked conformation aligns very well to the crystal bound conformation of resveratrol, Figure 4A. Second, binding of a quercetin molecule in S1 has improved the docking score of the quercetin molecule in S2 by 0.2 units to -8.6 which in turn has allowed a quercetin molecule to dock favorably in S3 with a docking score of -8.2, Table 2. Importantly, no favorable docking pose could be detected for quercetin in S3 with an unoccupied S2. This suggests the cooperative binding of quercetin to Sirt-1, Figure 4A, similar to resveratrol. Third, all three docked conformations of quercetin aligned well to their counterpart bound conformations of resveratrol in structure 5BTR, Figure 4A. Fourth, quercetin mediates H-bonding interactions between Sirt-1 and the peptide substrate similar to resveratrol. The H-bonds detected with quercetin in the three subsites are quercetin in S1: it should be noted that in this subsite, ring A of quercetin binds where resveratrol’s ring B binds and ring B of quercetin binds where resveratrol’s ring A binds, Figure 4B. Another difference is the two OH groups on quercetin’s ring B are ortho to each other whereas the two OH groups on resveratrol’s ring A are meta to each other. One of the two OH groups on ring B of quercetin H-bonds with the side chain of Sirt-1’s E230 (the same H-bond is observed with resveratrol as explained above). While the meta OH in resveratrol’s ring A H-bonds to the substrate’s backbone carbonyl oxygen of residue K3, the corresponding ortho OH on quercetin’s ring B is not seen to H-bond to anything. However, the OH group on quercetin’s ring C (which doesn’t have a counterpart in resveratrol) H-bonds to the substrate’s backbone carbonyl oxygen of residue K3; this makes up for the H-bond that the ortho OH could not form. While the sole OH on resveratrol’s ring B H-bonds to Sirt-1’s backbone amide nitrogen of P211, the two OH groups on quercetin’s ring A do not H-bond to Sirt-1 or its peptide substrate. Quercetin in S2: in this subsite, ring A of quercetin binds where ring A of resveratrol binds and ring B binds where ring B binds. With that being said, one of the OH groups on quercetin’s ring B maintains the same interactions between Sirt-1 and the substrate as the sole OH group of resveratrol’s ring B. That is, it H-bonds to the side chains of Sirt-1’s Q222 and N226 as well as to the substrate’s backbone carbonyl oxygen of R1. Also, the ortho OH group on quercetin’s same ring, B (which does not have a counterpart in resveratrol’s ring B), H-bonds to the substrate’s both K3 backbone amide nitrogen and R1 backbone carbonyl oxygen. This is a stabilizing interaction that is not observed with resveratrol. Also, ring C of quercetin (which also does not have a counterpart in resveratrol) has an OH group that H-bonds to the terminal amino group of the substrate’s K3 side chain; another stabilizing interaction that is not observed with resveratrol. Ring A of quercetin, which has the same meta OH groups of resveratrol, has one of its OH groups H-bonding to Sirt-1’s side chains of D298 (same as resveratrol’s ring A OH group), while the other OH H-bonds to the side chain of Sirt-1’s E300 (an H-bond that is not observed with resveratrol). Quercetin in S3: in this subsite, quercetin’s ring A binds where resveratrol’s ring B does. One of the meta OH groups on quercetin’s ring A H-bonds to the ring oxygen of the FDL fluorescent group on the peptide substrate (resveratrol’s ring B OH H-bonds to FDL’s carbonyl oxygen). Also, this same OH is seen to H-bond to Sirt-1’s K444 backbone carbonyl oxygen (same interaction is observed with resveratrol). The OH group on quercetin’s ring C is seen to H-bond to Sirt-1’s side chains of D292 (an interaction that is observed with resveratrol’s ring A OH) and Q294 (not observed with resveratrol). The two OH groups on quercetin’s ring B H-bond to the side chain of D298 (also observed with resveratrol’s ring A OH). It is to be noted that the quercetins in S1 and S3 are seen to form hydrophobic interactions with the fluorescent FDL group of the peptide substrate (similar to resveratrol). These docking results strongly suggest that quercetin is capable of mediating the same stabilizing interactions of resveratrol, if not more, which supports the likelihood that quercetin acts as a direct activator of Sirt-1 as was predicted based on the similar pharmacophoric features to resveratrol.
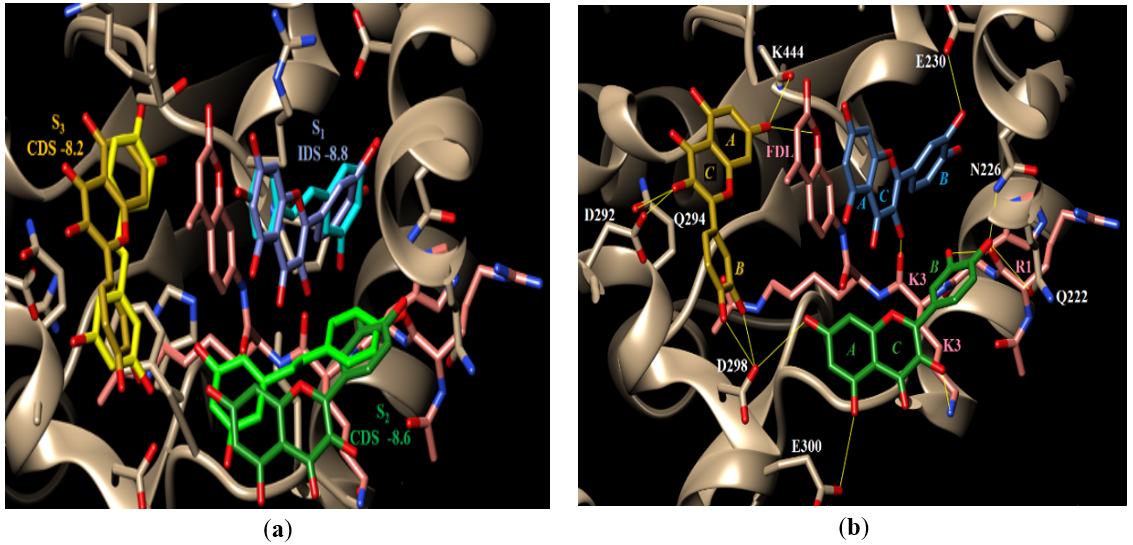 Figure 4. (a) Docking of quercetin. Shown are the docked conformations of quercetin in S1, S2, and S3 overlaid on the bound conformations of resveratrol in the same subsites in structure 5BTR. The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3) whereas quercetin molecules have darker shades of the same colors; (b) potential H-bonding pattern of quercetin.
Figure 4. (a) Docking of quercetin. Shown are the docked conformations of quercetin in S1, S2, and S3 overlaid on the bound conformations of resveratrol in the same subsites in structure 5BTR. The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3) whereas quercetin molecules have darker shades of the same colors; (b) potential H-bonding pattern of quercetin.
In addition to those resveratrol-like binding modes of quercetin, it also showed a crossover binding mode where one quercetin molecule bridged S1 and S2, Figure 5. Quercetin demonstrated five favorable (docking scores ranging from -9.3 to -9, Table 2) S1 – S2 crossover poses which were selected for further sequential docking. No favorable S2 - S3 crossover poses were observed.
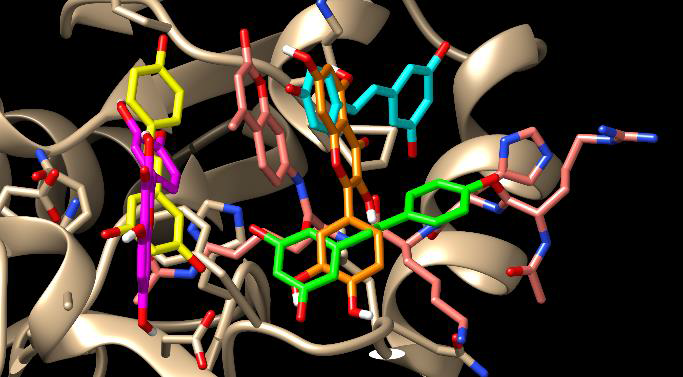
Figure 5. The S1-S2 crossover pose of quercetin. The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3). Quercetin molecules are shaded orange in the crossover S1-S2 site (IDS, -9.0) and pink in the S3 site (CDS, -8.5).
Sequential docking was completed with all five S1 – S2 crossover poses, each yielding favorable (docking scores ranging from -8.5 to -8.1) sequential binding in S3, Table 2. These crossover poses did not reproduce the pattern of hydrogen bonding observed with resveratrol and, therefore, were not considered biologically relevant. They are most likely an artifact of the rigidity of the quercetin and its larger size because of the additional ring. This rigidity makes it a little difficult for the molecule to fit in a smaller subsite and, instead, crosses over to a close-by subsite.
Table 2. Quercetin Docking Scores
|
Run |
Subsite |
IDS |
Run |
Subsite |
CDS |
Run |
Subsite |
CDS |
|
1 |
S1 |
-8.8 |
2 |
S2 |
-8.6 1 |
3 |
S3 |
-8.4 2 |
|
1 |
S2 |
-8.4 1 |
2 |
S3 |
-8.0 2 |
|
|
|
|
1 |
S1-S2 |
-9.03 |
2 |
S3 |
-8.5 |
|
|
|
|
1 |
S1-S2 |
-8.83 |
2 |
S3 |
-7.9 |
|
|
|
|
1 |
S1-S2 |
-8.83 |
2 |
S3 |
-8.4 |
|
|
|
|
1 |
S1-S2 |
-8.63 |
2 |
S3 |
-8.0 |
|
|
|
IDS and CDS of quercetin in the S1, S2, S3 sites as well as the S1-S2 and S3 combinations.
1demonstrated improvement of docking scores for S2 when S1 is previously docked.
2demonstrated improvement of docking score for S3 when S1 and S2 are already docked.
3crossover poses could not demonstrate improved docking as S3 was not occupied before the S1-S2 crossover site within this trial.
MHCP demonstrates alternate crossover poses and increased potential for hydrophobic interactions.
Comparing the structures of resveratrol and MHCP, it is clear that MHCP is not a polyphenolic compound, Figure 1. MHCP indeed has two aromatic rings but only one of them has a phenolic OH. The other ring carries only a methyl group, which severely limits its hydrogen bonding capacity. Another important structural difference between MHCP and resveratrol is the linker between the two aromatic rings. The linker in MHCP is one carbon longer than it is in resveratrol and it has a carbonyl group that does not exist in resveratrol. This longer linker in MHCP may not allow the molecule to fit conveniently in the Sirt-1 active site as resveratrol. Despite the limited H-bonding capacity and its larger size, MHCP was selected for docking due to its documented evidence in blood glucose control and speculated ability to function as an insulin mimic [25]. Also, it was hypothesized that the carbonyl group in MHCP linker may make up for the lack of other phenolic OH groups when H-bonding is considered.
Similar to resveratrol, MHCP docked most favorably into S1 with a docking score of -9.0. Unlike resveratrol, however, MHCP did not produce any biologically relevant docking poses in S2 that scored better than the -7.7 of the individual resveratrol molecule in the same S2. Docking a second MHCP molecule to this structure that contained the single S1 pose did not produce any biologically relevant poses in S2. Instead, the best scoring pose with this structure file was observed in S3 with a consecutive docking score (CSD) of -8.2, Figure 6(a). MHCP did not show the H-bonding pattern observed with resveratrol which is expected due to the lack of enough OH groups, Figure 6(b). MHCP in S1 is shown to H-bond to Sirt-1’s side chain of E230, Sirt-1’s backbone carbonyl oxygen of N226, and the substrate’s backbone carbonyl oxygen of K3. MHCP in S3 is shown to form only one H-bond with the Sirt-1 side chain of D298. This is a weak H-bonding pattern and, therefore, should not be expected to contribute much to stabilizing the substrate in the active site.
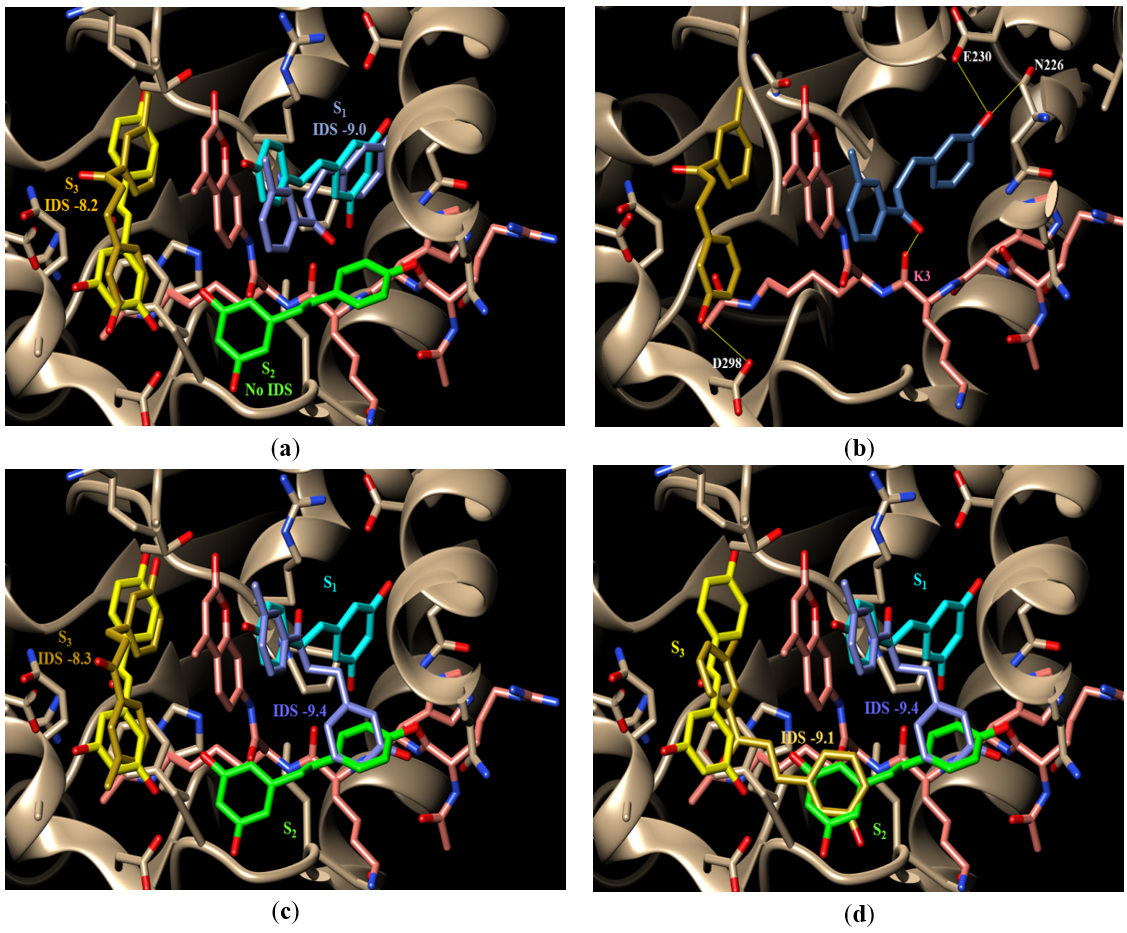
Figure 6. Docking of MHCP: (a) the docked conformations of MHCP in S1 and S3 overlaid on the bound conformations of resveratrol in the same subsites in structure 5BTR. The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3) whereas MHCP molecules have darker shades of the same colors; (b) the H-bonding pattern of MHCP; (c) the S1-S2 docked conformation of MHCP (darker blue color) and its corresponding S3 docked conformation (darker yellow color); (d) the S1-S2 and the S2-S3 docked conformations of MHCP (darker yellow color)
.
MHCP also showed a docked pose that bridged across S1 and S2 with an IDS of -9.4, Figure 6(c). Docking a second MHCP to this structure with the S1-S2 pose produced two docking poses; an individual pose that remained local to S3 with a CSD of -8.3, Figure 6(c), and a crossover pose that bridged S2 and S3 with a CSD of -9.1, Figure 6(d). The H-bonding pattern seen with those poses was weak (data are not shown). The phenolic OH of the S1-S2 crossover pose is seen to H-bond to Sirt-1’s side chain of Q222 and the substrate’s backbone carbonyl oxygen of R1 while the phenolic group of the local pose in S3 is seen to H-bond to Sirt-1’s carbonyl oxygen of K444 and the oxygen of the FDL fluorescent group. On the other hand, the S2-S3 crossover pose was not seen to form H-bonds. Again, this weak H-bonding pattern of MHCP is due to its limited number of H-bonding groups and, therefore, cannot be expected to contribute to substrate stabilization in the active site.
In general, MHCP was more proliferative than quercetin with many more crossover poses bridging S1- S2 than quercetin. This is most likely a reflection of MHCP’s structural flexibility (more rotatable bonds) and bigger size (longer linker) over quercetin and even resveratrol, which limits MHCP’s ability to fit into only one subsite. This is supported by the higher docking scores of the crossover poses over the localized ones, Table 3. MHCP docking poses suggested extremely limited ability to stabilize substrate via H- bonding as was observed with resveratrol and quercetin. Although docking scores form MHCP were favorable, we suspect they were inflated by the molecule’s flexibility and the hydrophobic interactions it forms.
Table 3. MHCP Docking Scores.
|
Run |
Subsite |
IDS |
Run |
Subsite |
CDS |
Run |
Subsite |
CDS |
|
1 |
S1 |
-9.0 |
2 |
S3 |
-9.0 |
3 |
S2 |
-7.5 |
|
|
|
|
2 |
S3 |
-8.2 |
|
|
|
|
1 |
S1-S2 |
-9.4 |
2 |
S2-S3 |
-9.1 |
|
|
|
|
|
|
|
2 |
S3 |
-8.9 |
|
|
|
|
1 |
S1-S2 |
-9.1 |
2 |
S3 |
-9.1 |
|
|
|
IDS and CDS of MHCP compounds in S1, S2, and S3 and crossover poses.
Cinnamic acid groups of molecules exhibit unfavorable docking scores.
The molecules in this group include cinnamic acid, cinnamyl alcohol, and cinnamaldehyde which, together, represent the major component of cinnamon extract (65%-80%) [5]. Each of those molecules was docked into the evacuated Sirt-1 active site. Similar to resveratrol, quercetin, and MHCP, their best scoring docking poses were in S1 with docking scores of -7.2 for cinnamic acid, -6.8 for cinnamyl alcohol, and -6.7 cinnamaldehyde; Table 4.
Despite this fact, none of the poses obtained with this group of molecules were considered biologically relevant because 1) none of the obtained poses matched the S1 bound conformation of resveratrol in structure 5BTR, 2) these docking scores are significantly less than the baseline values calculated for resveratrol in S1, and 3) upon sequential docking, the subsequent molecules docked outside the allosteric binding site. These results are not surprising because, unlike resveratrol, this group of molecules only have one aromatic ring and it does not carry any phenolic OH groups. For all those reasons, we believe that it is highly unlikely that any of the molecules in this group is a direct activator of Sirt-1.
Table 4. Cinnamic Acid Compounds Docking Scores.
|
Compound Name |
Most Favorable Docking Score |
|
Cinnamic acid |
-7.2 |
|
Cinnamyl alcohol |
-6.8 |
|
Cinnamaldehyde |
-6.7 |
IDS and CDS of cinnamic acid compounds in S1, S2, and S3.
Eugenol demonstrates poor docking scores indicating it is not likely a Sirt-1 activator.
The docking results for eugenol were very similar to those obtained with the cinnamic acid group of molecules. It preferentially docks in S1 with a score of -6.8 for the best scoring pose but none of the poses obtained were considered biologically relevant for similar reasons to the cinnamic acid group (data are not shown). Although eugenol has one phenolic OH group, its size and chemical structure are more similar to the cinnamic acid group than it is to resveratrol. These results, therefore, are only to be expected.
Select Catechin isomers show potential for Sirt-1 activation, with (+)-epicatechin (2S,3S) exhibiting the most favorable docking scores.
Although not all four isomers of catechin are as prevalent in the cinnamon extract, many natural products do contain a mixture of all four isomers. For this reason, all four isomers were examined for docking potential. Two of the four catechin isomers docked, (-)-catechin (2S,3R), and (+)-epicatechin (2S,3S), showed favorable scores. The two R isomers, (-)-epicatechin (2R,3R), and (+)-catechin (2R,3S), although potentially biologically active were excluded because docking scores exceeded our assigned baseline of -8. Sequential docking was completed with the two favorable isomers, (-)-catechin (2S,3R) and (+)-epicatechin (2S,3S), Figure 7. Scores for docking of the favorable two isomers are shown in tables 5.1 and 5.2.
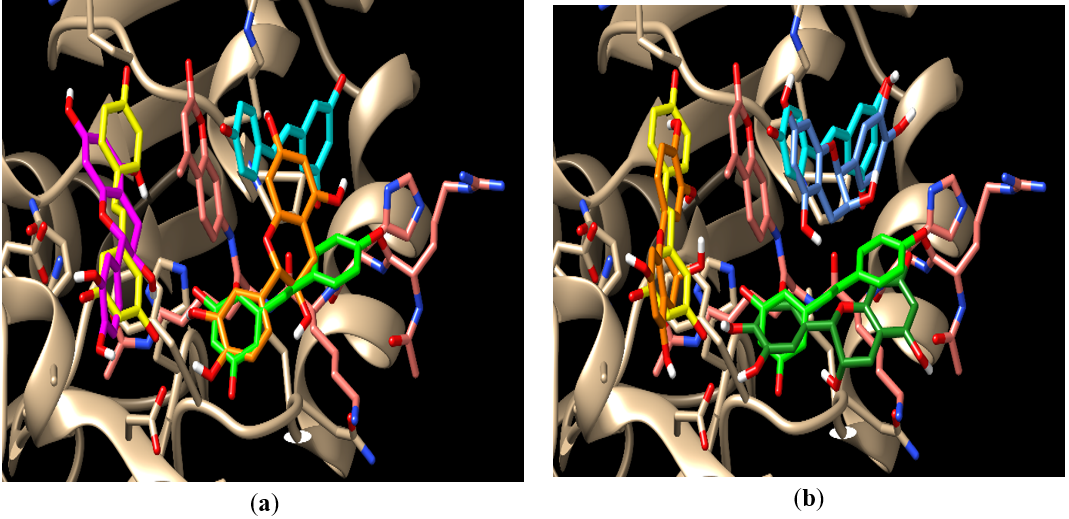 Figure 7. The docking poses for (a) (-)-catechin (2S,3R) and (b) (+)-epicatechin (2S,3S). The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3); (a) (-)-catechin (2S,3R) (orange) is shown docking in the S1-S2 crossover pose with an IDS of -8.9. The second molecule (magenta) is shown in S3 with a CDS or -8.2; (b) shows (+)-epicatechin (2S,3S) docking in S1 (dark blue), S2 (dark green) and S3 (orange). This simulation represents the most favorable docking scores for the (+)-epicatechin (2S,3S).
Figure 7. The docking poses for (a) (-)-catechin (2S,3R) and (b) (+)-epicatechin (2S,3S). The resveratrol molecules are colored light blue (S1), light green (S2), and light yellow (S3); (a) (-)-catechin (2S,3R) (orange) is shown docking in the S1-S2 crossover pose with an IDS of -8.9. The second molecule (magenta) is shown in S3 with a CDS or -8.2; (b) shows (+)-epicatechin (2S,3S) docking in S1 (dark blue), S2 (dark green) and S3 (orange). This simulation represents the most favorable docking scores for the (+)-epicatechin (2S,3S).
Table 5.1. (-)-catechin (2S, 3R) Docking Scores.
|
Run |
Subsite |
IDS |
Run |
Subsite |
CDS |
|
1 |
S1 |
-9.0 |
2 |
S3 |
-9.0 |
|
|
|
|
2 |
S3 |
-8.2 |
Table 5.2. (+)-epicatechin (2S,3S) Docking Scores.
|
Run |
Subsite |
IDS |
Run |
Subsite |
CDS |
Run |
Subsite |
CDS |
|
1 |
S1 |
-9.0 |
2 |
S2 |
-8.7 |
3 |
S3 |
-8.4 |
|
1 |
S1-S2 |
-8.8 |
2 |
S3 |
-7.9 |
|
|
|
IDS and CDS of catechin 2S3S and 2S3R in the three subsites S1, S2, and S3.
The two S-isomers also showed cooperative binding in the sequential docking simulations. The (+)-epicatechin (2S,3S) isomer exhibited the most favorable sequential docking scores with -9 in S1, -8.7 in S2, and -8.4 in S3. These favorable scores suggest, that although minimal hydrogen bonding exists in these sequential docking trials with (+)-epicatechin (2S,3S), there is potential for this isomer to function as a Sirt-1 activator potentially via hydrophobic interactions.
CONCLUSIONS
Many naturally occurring polyphenolic molecules are biologically active. Resveratrol has gained much interest due to its suggested health benefits, in particular its anti-hyperglycemic effects. Research has demonstrated that Sirtuin enzymes play an active role in mediating the acetylation status of genes involved in metabolism, aging, and apoptosis [26-28]. Research has shown that these health benefits are due to the activation of Sirt-1.
Experimental results have shown direct binding and activation of Sirt-1 by resveratrol. Development of a reliable and relatively rapid screen for molecules with the potential for activating Sirt-1 and mediating these deacetylation reactions, therefore, would allow researchers to better predict which molecules have the potential to be used therapeutically, thus warrants more aggressive assay of Sirt-1 activities. Resveratrol is a polyphenol that bears structural similarities to many components in the cinnamon extract. Our docking results suggest a method to identify components of cinnamon extract that have the potential to function as a Sirt-1 activator.
Our docking protocol with resveratrol has reproduced the experimentally observed binding modes with resveratrol in structure 5BTR with great accuracy indicating the validity of our method. We then investigated the docking behavior of the different components in the cinnamon extract to Sirt-1. Quercetin, the major polyphenolic component in the cinnamon extract, displayed docking behavior and an H-bonding pattern that was very similar to those observed with resveratrol; strongly suggesting that it can stabilize the substrate in the active site and act as a Sirt-1 activator similar to resveratrol.
Catechin is a structurally similar molecule to quercetin. A major structural difference, however, between the two molecules is the lack of planarity of catechin due to saturation of the double bond and loss of the carbonyl oxygen from the oxygen-containing ring, Figure 1. This lack of planarity of catechin prevents the molecules from fitting properly in the different subsites which makes it difficult for the molecule to form the same H-bonding pattern seen with quercetin and observed with resveratrol. Catechin, therefore, may not be an efficient stabilizer of the substrate in the binding site and, as a result, may not be a good activator of Sirt-1 according to our model
MHCP is similar in structure to resveratrol with two major difference; 1) it only has one phenolic OH (resveratrol has three and quercetin has five), and 2) a three-carbon linker (two-carbon linker in resveratrol) between the two aromatic rings that also contains a ketone group (not present in resveratrol linker). Docking of MHCP into the Sirt-1 active site produced many of the crossovers poses more than quercetin but none of those poses was seen to contribute significantly to substrate stabilization in the binding site. The bigger size of MHCP did not allow it to fit properly in one small subsite and, instead, it frequently crossed over to other subsites. Also, the lack of so many OH groups did not allow the molecule to bridge between Sirt-1 and its substrate peptide through the pattern of H-bonding observed with resveratrol. For these two reasons, it is highly unlikely that MHCP would be able to stabilize the substrate in the binding site or act as a Sirt-1 activator. The docking scores obtained with MHCP may be “favorable” but we believe they are inflated as a result of its flexibility (more rotatable bonds than resveratrol) and the hydrophobic interactions it forms in the binding site.
If both catechin and MHCP have been reported to have anti-hyperglycemic effects experimentally, it could be due to either 1) binding in an allosteric site outside of the binding site where resveratrol binds, or 2) working through a different mechanism that does not involve Sirt-1 activation.
The other components in cinnamon (the cinnamic acid group and eugenol) did not show a strong affinity to Sirt-1 nor did they form the hydrogen bonding pattern seen with resveratrol. However, these other cinnamon components have been reported to have anti-hyperglycemic effects as well. Again, this most likely suggests that they act on an alternative target to achieve those anti-hyperglycemic effects. It is also possible that these molecules activate Sirt-1 via a mechanism unrelated to the one proposed in this docking study.
The docking method we presented here was limited to components of cinnamon extract but the method can be extrapolated to include other natural products and synthetic molecules that have the potential to function as a Sirt-1 activator. Also, our docking protocol allowed us to predict the sequential and cooperative binding of multiple molecules to stabilize a relatively large allosteric site with the potential to alter Sirt-1 activity.
MATERIALS AND METHODS
Computer Specifications.
All docking calculations were performed on 64-bit Microsoft Surface with an Intel® Core™ i5-6300U CUP @ 2.40GHz and an 8.00 GB RAM.
Software Used in Preparation, Simulation, and Visualization.
MAESTRO [29] was used to build and optimize ligand structures. AUTODOCK VINA (VINA) [30] was used for all docking calculations performed in this study. CHIMERA [31] was used to visualize and analyze the docking results via the “viewdock” facility.
Protein structure preparation
The structure of the human Sirt-1 deacetylase enzyme in complex with the P53 substrate peptide and three resveratrol molecules (PDB code 5BTR [9]) was used for all the docking studies performed here. To prepare the structure for docking, the substrate peptide was kept in place while the three resveratrol molecules were manually removed from the receptor PDB file. The structure file of the receptor was then imported to AUTODOCK MGL TOOLS 1.5.6 (ADT) [32, 33] to define the binding site docking box and generate the pdbqt receptor file for the docking engine. To do that, atoms were assigned an “autodock type” and only polar hydrogens were added. Second, non-polar hydrogens were merged to their respective carbons and the atomic Gasteiger charges [34] were assigned. To define the binding site for VINA, a grid box centered on the P53 peptide and made big enough to include all atoms of the binding site residues. The center of the box and the box dimensions are shown in Table 6.
Table 6. Docking box center coordinates and dimensions.
|
Box Center Coordinates |
Box Dimensions |
|
center_x = -17.173 A |
size_x = 25.3 A |
|
center_y = 68.970 A |
size_y = 31.878 A |
|
center_z = 15.701 A |
size_z = 25.3 |
Ligand preparation
The structures of resveratrol and all other molecules docked in this study were built and the geometries were optimized using MAESTRO. The optimized ligand structures were saved in the PDB file format to be imported into ADT where rotatable torsions were defined, atoms were assigned an “autodock type” and only polar hydrogens were added. Non-polar hydrogens were merged to their respective carbons and the atomic Gasteiger charges were assigned.
Other docking parameters
Gridpoint spacing was set to 1Å and a total of 20 docking poses were generated for each ligand with the exhaustiveness parameter set to 20 and the num_modes variable was set to 200.
REFERENCES