



|
Bacteriological Analysis with Antimicrobial Sensitivity and Resistance Pattern in Blood Culture of Septicemic Patient from Different Wards of a Tertiary Care Hospital in India Rohit Tiwari, Suraj Sandil, Parminder Nain*, Jaspreet Kaur |
|
Department of Pharmacy Practice, M.M. College of Pharmacy, Maharishi Markandeshwar (Deemed to be University) Mullana-Ambala (Haryana) India. |
ABSTRACT
Background: Septicemia is one of the leading causes of morbidity and mortality in India where a large number of broad-spectrum antibiotics as empirical therapy implement against causative microorganism has led to the emergence of multidrug-resistant of these strains. Object: The present study was conducted to determine bacterial etiology with antibiotic susceptibility and resistance profile of isolated organisms from septicemia. Methods: This retrospective observational study was conducted over a period of six months to evaluate the blood culture, sensitivity, and resistance pattern of various antimicrobial agents used in septicemic patients. Results: We collected and analyzed a total of 380 blood culture reports during the study period of which 345 (90.78%) samples were found to be positive, out of which 62.02 % was found to be gram-positive bacteria whereas 37.98% was gram-negative bacteria. The most commonly isolated organisms were Staphylococcus aureus (83.28%), Micrococcus species (6%), Enterococcus species (6%), and Staphylococcus saprophyticus (5%) under gram-positive bacteria. The majority of organisms isolated were resistant to commonly used antibiotics like 2nd & 3rd generation cephalosporin, which showed more than 90% resistance for gram-positive and gram-negative organisms. Conclusion: The results obtained from this study showed aminoglycosides as a highly sensitive antibiotic and cephalosporin as a highly resistant antibiotic. This study would guide the clinicians to formulate appropriate treatment strategies as well as to take various preventive measures which ultimately would help to decrease sepsis-related mortality.
Key Words: Septicemic, Antimicrobial sensitivity/resistance, Bacteriological analysis.
INTRODUCTION
Infectious diseases have always constituted the most serious health issue in the world [1-3]. Septicemia also known as bacteremia or blood poisoning [4], also known as bacteremia or blood poisoning, is a life-threatening condition with multiple complications of blood which is very serious in nature [5]. It is usually the result of untreated or severe bacterial infection which is present elsewhere in the body parts such as lungs and skin, enters the bloodstream [6]. The causative organism is transmitted through the bloodstream to the whole body, which leads to multiple organ failure basically heart and brain [7]. The specific source cannot be determined by its etiology but multiple infections further could lead to septicemia if remained untreated, like urinary tract infection, abdominal infection, lung infection, or any other infection [8]. There are many causative organisms responsible for the disease; they may be gram-positive, negative, or anaerobic bacteria depending upon the preceding infection. Patients already in the hospital are at a higher risk of developing septicemia, and the hospital stay also increases the chances of mortality. The blood infections occurring by hospital sources are mostly more dangerous as compared to other sources because these causative organisms are resistant to multiple antibiotics at higher doses. Sepsis and septic shock is the most common complication of septicemia [9]. It produces inflammation throughout the body that causes blood clots and prevents oxygen from reaching vital organs, resulting in organ failure. The incidence rate of severe sepsis in the United States is estimated to be 300 cases per 100 000 population [10].
Septicemia is a medical emergency that can be treated effectively with antibiotics. Initial treatment is performed with broad-spectrum antibiotics as “empirical therapy” without figuring out the type of bacteria but antibiotic resistance has become a burning issue worldwide leading to an increase in death of the patients with septicemia [11]. Having control over the infection has become a very difficult task especially in developing countries like India where infection still is a major cause of death [12]. There are several factors like point mutations, gene amplification, and extrinsic factor i.e. horizontal transfer of the resistant gene between bacteria, and within species by transposons, integrin’s or plasmids are the proposed postulate for the resistance development in bacteria [13]. It cannot be reduced the irreversible change either developed by limiting antibiotic use. Overcrowding, deficient hygiene, and demographic changes are some of the social factors for the outcome of antimicrobial resistance which have been supported by multidrug resistance [14]. According to the WHO (world health organization), the inappropriate and irrational uses of antibiotics are noted to be the major cause of hospital and community-acquired infections [15]. As antibiotic sensitivity and resistance pattern to common pathogen has been changing day by day, so there is the need for constant antimicrobial sensitivity surveillance. Common culture study and their antibiotic sensitivity/resistance pattern help to set the rational and correct use of antibiotics in the regions [16]. It will support the clinicians in providing effective and safe, empirical therapies, develop a rational prescription pattern, and finally make policy decisions. Assessment of antibiotic sensitivity patterns in periodic intervals is mandatory for choosing appropriate antibiotic therapy in each region [17]. Hence, it has been necessary to study bacteriological analysis and antibiotic resistance or sensitivity pattern. The present study was undertaken to study the bacteriological profile of septicemia cases and their antibiotic susceptibility pattern for planning strategy for the management of these cases.
METHODOLOGY
Study Design
A retrospective record-based study was conducted in the microbiology department for six months from 1 July 2018 to 31 December 2018 at a tertiary care hospital located in the northern part of India. We took data of adult patients from both sexes. The study was done only on the basis of blood culture and sensitivity reports in the microbiology department. There was no direct involvement of the patient or any other information related to the case, diagnosis, and other investigations. Various microbiological cultures and testing were done as per standard guidelines. The study was conducted after due approval from the institutional review committee.
Sample Size and Study Population
We included 345 positive culture reports of adult patients from different wards like RICU, MICU, SICU, ICU, and their respective wards, who were suspected/diagnosed cases of septicemia and in whom blood cultures were done.
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was done for the isolates on Muller-Hinton agar using commercially available discs (Hi-Media) by Kirby-Bauer disc diffusion method, using the Clinical and Laboratory Standards Institute (CLSI) guidelines for interpretation. Every batch of Mueller-Hilton agar and antibiotic discs were tested by using the American Type Culture Collection (ATCC) control strains.
Statistical Analysis
The data was analyzed by using Microsoft Excel sheet and expressed in percentage using descriptive statistics and pie charts.
RESULT
In this study, out of 380 cases samples received by the microbiology department for blood culture investigation with antibiotic resistance and sensitivity reporting, 345 were positive and documented for further analysis.
The total positive cases reported were considered for the result that showed the ratio of males and females was 1:1 i.e. 50% female and 50% male (Figure 1).
Bacteriological Analysis
After analysis, the data showed that about 345 (90.78%) blood cultures were positive for septicemia, confirmed by growth on the culture plate, of which 214 (62.02%) were positive for gram-positive bacteria and 37.98% for gram-negative bacteria (Figure 1). In the present study, gram-negative organisms constituted the major group of isolates from septicemia cases.
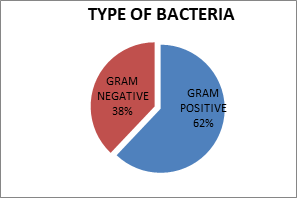
Fig. 1. Distribution of bacteria in blood culture reports of the patients.
(Gram-positive and gram-negative bacteria).
The isolation pattern of Gram-positive organisms showed the most frequently isolated organisms were Staphylococcus aureus (83.28%), Micrococcus species (6%), Enterococcus species (6%), and Staphylococcus saprophyticus (5%) (Figure 2).
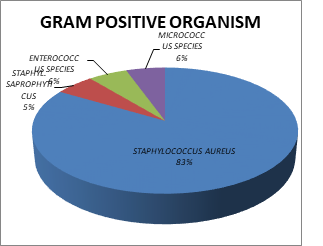
Fig. 2. Percentage of Gram-positive organisms in blood culture of patients
The isolation pattern of gram-positive organisms showed the most frequently isolated organisms were Citrobacter Species (28%), Acinetobacter Species (27%), Pseudomonas species (9%), Escherichia coli (9%), Pseudomonas aeruginosa (9%), and Enterobacter species (9%) (Figure 3).
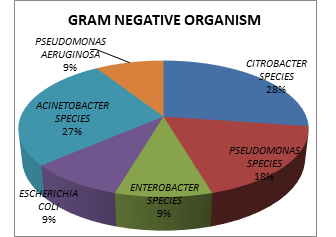
Fig. 3. Percentage of Gram-negative organisms in blood culture of patients.
Antibiogram against Gram-Positive microorganisms: The results of antibiotic sensitivity/resistance pattern (Table 1) revealed that Staphylococcus aureus organism was resistant to the majority of antibiotics. The maximum resistance (more than 80%) was observed against Penicillin (amoxicillin and other penicillin); other antibiotics that showed more than 50% cases of resistance were aminoglycosides (azithromycin, erythromycin), Fluoroquinolones (ciprofloxacin, levofloxacin), and Sulfonamides (co-trimoxazole). Isolated Staphylococcus aureus was sensitive to Glycopeptide (vancomycin) (92.85%), gentamycin (87.5%), linezolid (85.71%). meropenem (86.19%), and chloramphenicol (73.33%) (Figure 4).
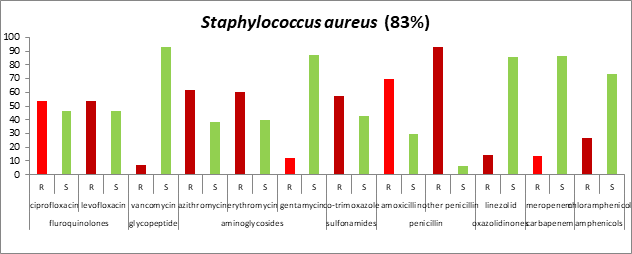
Fig. 4. Antibiogram against Gram-positive (Staphylococcus aureus) microorganisms
Antibiogram against Gram-negative microorganisms: Most of the Gram-negative organisms were resistant to commonly used antibiotics (Table 1). Isolated. Citrobacter species showed maximum resistance against amoxicillin (75%) followed by co-trimoxazole, gentamycin, chloramphenicol, and ciprofloxacin. This species was one hundred percent (100%) sensitive to levofloxacin, azithromycin, and meropenem (Figure 5).
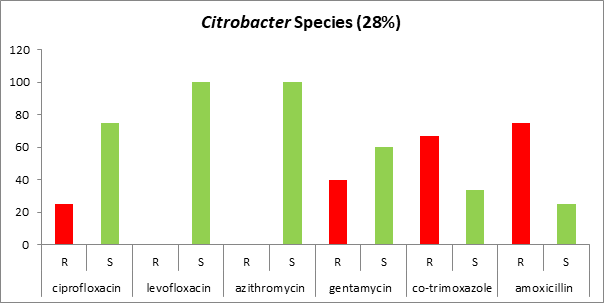
Fig. 5. Antibiogram against Gram-negative (Citrobacter Species) microorganisms.
Isolated Acinetobacter species showed more resistance than other isolated gram-negative bacilli; 100% of them were resistant to gentamycin and amoxicillin followed by 75% resistance to co-trimoxazole and 50% to ciprofloxacin and chloramphenicol. This species was 100% sensitive only to levofloxacin (Figure 6).
Fig. 6. Antibiogram against Gram-negative (Acinetobacter Species) microorganisms
All isolated Pseudomonas species were resistant against amoxicillin (100%), followed by co-trimoxazole (83%), levofloxacin (60%), ciprofloxacin (33%), and gentamycin (20%), while they were sensitive to many of antibiotics like 100% sensitive to chloramphenicol and meropenem followed by 80% sensitive to gentamycin, 66% sensitive to ciprofloxacin, and 40% sensitive to levofloxacin (Figure 7).
Fig. 7. Antibiogram against Gram-negative (Pseudomonas Species) microorganisms.
DISCUSSION
Septicemia is one of the major causes of morbidity and mortality across India. For the effective management of septicemia, the study of the bacteriological analysis with their antibiotic biogram plays an important role [18]. In our study, a total of 380 microbial culture and sensitivity tests were done during the study period, of which 90% were found to be positive. In our hospital, culture and sensitivity reports are done on the day of admission, and empirical antimicrobial therapy is started on the same day. In this study, gram-positive organisms were identified in 62% of cultures and gram-negative organisms were seen in 38 % of cultures. This is higher than that reported by Gupta et al., Kamga et al. and Anbumani et al. who reported similar (gram-positive organisms) incidences were higher, but some workers such as Mehta et al., Mehdinejad et al. and Barati et al. have reported that gram-negative organisms as the predominant pathogen in hospital settings [19-24]. The variations in the prevalence of organisms in different institutes worldwide may be due to epidemiological differences of etiological agents, the inclusion criteria for selection of subjects, prior use of antibiotics, the clinical setting of the study, and presence or absence of risk factors.
In the present study, gram-positive organisms constituted the major and commonest group of isolates from septicemia cases. Among this group, staphylococcus aureus and in gram-negative organisms Citrobacter species and Acinetobacter species have been found to be the prominent pathogens, which correlates with the past findings [25]. The prescribing of antibiotics in septicemic patients is usually empiric. The results of the antibiotic sensitivity pattern revealed that the majority of gram-positive and gram-negative organisms were resistant to the commonly used antibiotics. In our study, we found that Staphylococcus aureus was resistant to penicillin but highly sensitive to glycopeptide, carbapenem aminoglycosides, and amphenicols class of antibiotics. This suggests that vancomycin, meropenem, gentamycin, and chloramphenicol are the most effective drugs for Staphylococcus aureus infections. The present study showed that gram-negative isolates were frequently resistant to gentamycin, co-trimoxazole, and ciprofloxacin, hence, they must be judiciously used to minimize the morbidity and mortality. Levofloxacin showed 100% sensitivity for some gram-negative organisms.
The result of this study showed the antibiotic of the same class was found to be sensitive and resistant in another patient. Bacterial strains resistant to most classes of antibiotics will emerge unless inappropriate utilization of drugs is curtailed and continuous education of infection control practice is performed. For the empiric management of infection, there is a necessity for continuous surveillance patterns and efficient hospital infection control. Thus, the ongoing surveillance of antibiotic susceptibility patterns of predominant bacteria is a fundamental effort to monitor modifications in susceptibility patterns and to help the clinician in selecting empirical or directing therapy appropriately and also in preventing the emergence of multi drug-resistant bacteria.
CONCLUSION
Septicemia is a life-threatening emergency, and fast treatment with antibiotics is critical for a favorable outcome. This study brings us to the conclusion that being a microbiologist, our duty is not only to identify the organism and report about its sensitivity but also to guide the clinicians about the proper preventive measures to be taken. By this study, we want to highlight the fact that Gram-positive organisms, especially Staphylococcus aureus are the predominant group of organisms causing hospital-acquired septicemia in our institution and were found to be resistant against penicillin, thus revealing that the use of this class of drugs might be ineffective. Thus, great caution is needed for the selection of antibiotic therapy. Resistance to antibiotics poses a serious and growing issue, as such resistant bacteria are becoming more difficult to treat.
This reveals the variable nature of antibiotic susceptibility patterns, both in time and location. Therefore, it is advisable to continuously assess the sensitivity-resistance pattern of isolates to make rational utilization of antibiotics. These data support the hypothesis that the determination of antibiotic sensitivity patterns in periodic intervals is mandatory in each region for choosing appropriate antibiotic therapy. In the view from above, the strategy of antibiotic usage in the hospital must be reviewed in a laboratory that would prove a great help for the clinicians. The study might be helpful for the analysis and change in therapy for better outcome of the therapy according to the changing bacterial resistivity in the hospital and north India population.
Limitations of the study
Antibiotic disc sensitivity test results may vary with the hospital setting, while the infection rate in a hospital may depend on the hospital environment, antibiotic use, and other infection control practices. All these would limit the applicability of the findings of this study to other hospital settings.
ACKNOWLEDGMENT
We are very much thankful to the Head of the department of microbiology and staff of the microbiology laboratory for their encouragement and needful support. No financial support was taken for this study.
Conflict of interest
We, the authors of this article, declare that we have no conflict of interest of any means.
REFERENCES